Class 21: De Bruijn Assembly
Bioinformatics
Andrés Aravena
23 December 2021
Assembly statistics
Assembly Statistics
When we report an assembly result, we describe
Depth of coverage
Breadth of coverage
Number of contigs
N50
Two reads connect when they overlap
We only see overlaps over a threshold \(T\)
The best \(T\) has to be big enough to guarantee that the reads do not overlap by chance
Bigger values of \(T\) reduce the probability of “overlap by chance”
What is the overlap size?

We can simulate in the computer
Negative overlap are gaps

If two reads overlap…
…they are in the same Contig
A group of contiguous sequences is called Contig
The goal of the assembly process is to find one contig.
The sequence of this contig will be the genome
Most of times we do get several contigs
Num. contigs depends on 𝐺, 𝐿, 𝑁 and 𝑇

- 𝐿 =100
- 𝐺 =106
- 𝑇 =20
How many reads shall you pay?
What if reads are longer?

- 𝐿 =300
- 𝐺 =106
- 𝑇 =20
Longer reads are better
This is described in the paper
Lander ES, Waterman MS. Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics. 1988 Apr; 2(3):231-9. doi: 10.1016/0888-7543(88)90007-9. PMID: 3294162.

Contig length
With this simulation we can also calculate the length of each contig.

N50
Given a set of contigs, the N50 is defined as the sequence length of the shortest contig at 50% of the total genome length
We sort the contigs from largest to smallest

When do we get 50%?

We identify which contig crosses the 50% line
N50 is the length of that contig
Which contig reach 50% of bases?
| sorted_len | pct_in_contig | cumulative |
|---|---|---|
| 1981 | 30.40 | 30.40 |
| 1202 | 18.44 | 48.84 |
| 1055 | 16.19 | 65.03 |
| 677 | 10.39 | 75.42 |
| 581 | 8.92 | 84.33 |
| 511 | 7.84 | 92.17 |
| 510 | 7.83 | 100.00 |
Assembly quality
Each fragment has two reads
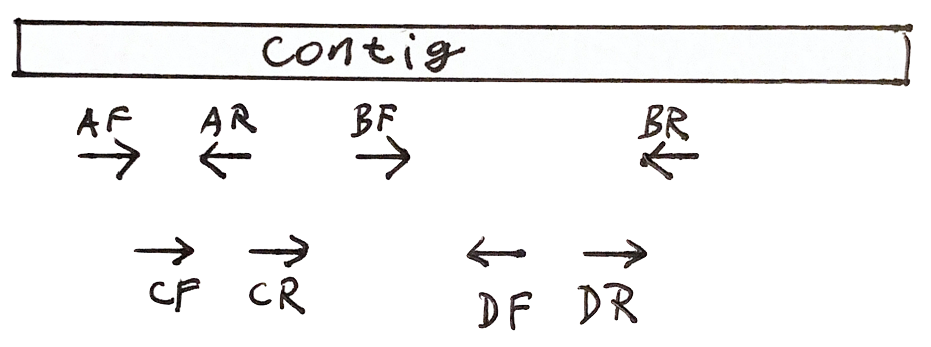
- Both reads should point to each other (AF, AR)
- If they point in the wrong direction, it is bad
- The distance between both reads should not be too large or too small
Scaffolding
If a fragment has one read in Contig 1 and the other in Contig 2, then we know that the contigs are close
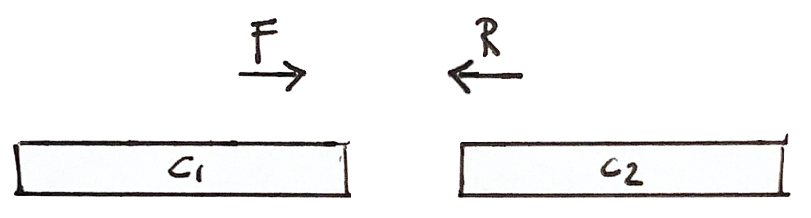
Scaffolding
More shared fragments gives more confidence to the scaffold
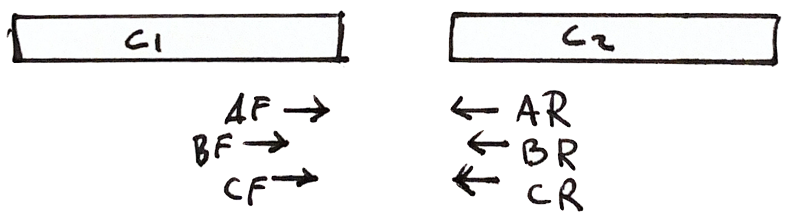
Scaffolding
Shared fragments allow us to find the relative orientation of contigs, and make a scaffold of contigs and gaps 
The problem of Repeats
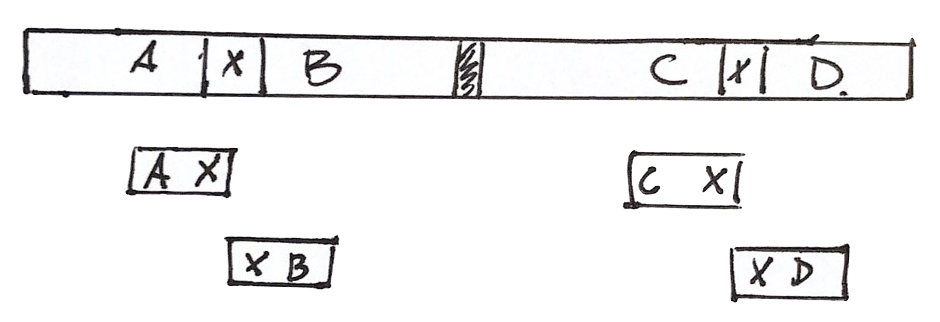
The assembly can be wrong
Given the information we have, there are at least two solutions

We cannot do better with the available information
We need more information
To decide the correct one, we need more information
For example, longer reads containing the repeat and its context

Or read pairs from larger fragments, so each read is outside the repeat
Another way to assemble
The problem
Assemblers based on Overlay–Layout–Consensus cannot handle repeats
Moreover, they can handle only a few thousand reads
NGS produces millions of reads, so a different approach was developed
𝑘-mers instead of reads
Since a read has hundreds of bp, it can contain part of a repeat
It is better to use shorter sequences (𝑘-mers), that are either inside or outside the repeat
𝑘 is typically between 20 and 130, and can be chosen automatically
Each read is translated into a list of 𝑘-mers
ATGCATATATAGCA
ATG
TGC
GCA
CAT
ATA
TAT
ATA
TAT
ATA
TAG
AGC
GCAWe do not count repeated 𝑘-mers
ATGCATATATAGCA
ATG
TGC
GCA
CAT
ATA
TAT
TAG
AGC
GCADe Bruijn’s graphs
Each 𝑘-mer is a node in a directed graph
Two nodes are connected if the last (𝑘-1) bp of the first 𝑘-mer are the same as the first (𝑘-1) bp of the second one
ATGCATATATAGCA
ATG TGC GCA CAT ATA TAT TAG AGC GCAWe get an honest assembly
This approach does not solve the repeats
Instead, it shows the repeats clearly

This way we know what are the issues, and we can design an experiment (PCR?) to solve them
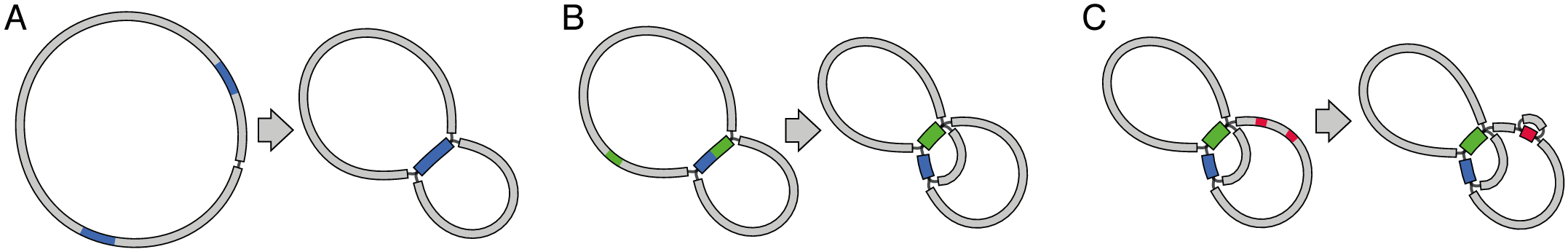
Repeats result in a funny graph
Output format: FASTG
Like FAST, but showing also the graph structure. For example
>EDGE_641517_length_474_cov_1.855908;
AACACTGATTGCCTCCCCCCCGTTGATGGGTAAAATAGCCGCAATTTTTCGTTTTCAACA
[…]
GCTGCCTGATGGTTATCGACGCTGCAAAAGGTGTTGAAGATCGTACCCGTAAGC
>EDGE_621787_length_514_cov_1.860465';
TGTCGATGCGGTGTACATTGTGGCAACGCCGGGTGAAATCGCTTTTATCAAACCGATGAT
[…]
TGGCTGGAAGGCAAAGGACTGCGGTTTATCGCCG
>EDGE_678376_length_822_cov_333.633094:EDGE_679076_length_4092_cov_132.576797',EDGE_679634_length_28752_cov_122.881432;
GGCACTGTTGCAAATAGTCGGTGGTGATAAACTTATCATCCCCTTTTGCTGATGGAGCTG
[…]
AGACAAAAGGCTGCCTCATCGCTAACTTTGCAACAGTGCCGGBandage: Look and edit the graph
Bandage is a program to visualize a assembly graph
Example from The New York Times: “Team of Rival Scientists Comes Together to Fight Zika”. March 30, 2016

Online Material
Indiana University Bloomington, “Introduction to Bioinformatics”, Lecture by Yuzhen Ye
Université Lyon I, Network Algorithms for Molecular Biology lesson on “Introduction to (de novo) assembly”, by Blerina Sinaimeri
MIT, “Foundations of Computational Systems Biology”, Lecture by David K. Gifford