Class 1: Why? How?
Methodology of Scientific Research
Andrés Aravena, PhD
17 March 2021
Welcome
to MSR 🌽
Today’s questions
Who
Why
How
What
Who?
I am Andres Aravena
- Assistant Professor at the Molecular Biology and Genomics Department
- Mathematical Engineer, U. of Chile
- PhD Informatics, U Rennes 1, France
- PhD Mathematical Modeling, U. of Chile
- not a Biologist
- but an Applied Mathematician who can speak “biologist language”
Why?
Why are you here?
Answer now with your voice
What?
Gene expression
In this course we will speak about
Transcription
We will learn to analize gene expression, so we can design better experiments and achieve higher impact
How does this fit in the Big Picture of Science
What is Science
Since there is no “authority”, nobody can make an “official” definition of Science.
There are two ways to “define” Science:
- Science is what scientist do
- very much used in practice
- but kind of useless definition
- does not allow to separate Science from PseudoScience
- Science is the result of applying the scientific method
An operational definition of Science
Scientist work is to understand Nature
We start by Observing Nature, usually measuring values.
These are exploratory experiments.
We study this in other courses.
The thing we study must be repetible, and we need to see that repetition.
We can find them using plots, linear models, clustering, etc.
This is the most important part.
Good answers to bad questions are useless.
Good questions are good, even if we don’t have answers
We answer these questions using models and explanations
Valid models should make predictions that we can test in the lab…
These are validation experiments.
If the results do not match the prediction, we know that the explanation is wrong. Two steps back.
Now we publish our data and model, so other scientists validate or reject it.
The final validation is to be published.
If the paper is accepted and published, our work becomes part of our shared human knowledge.
The goal of Science is to produce new Knowledge.
When we observe Nature we use our previous Knowledge
We look for new Patterns that raise new Questions.
“Noise becomes Signal”
Details
- Our Observations depend on our previous Knowledge
- The first step is to Find Patterns
- The key is to ask Good Questions
- Explanations are “models”, in a broad sense
- Valid models should produce new Predictions
- Observations and Test can be done in the lab
- “Knowledge” means Published
Characterization of Science
- About “outside”
- About visible things
- Things that you can measure
- Provides Explanations
- They must be Logic and Coherent
- Peer reviewed
- Replicable
Assumptions of Science
- There is an “objective reality” outside us
- The reality has some rules
- It is not (completely) random. There are rules
- The rules are “logic”
- The rules do not change
- We may not see the rules directly
- We can (in theory) discover these rules using reason
- Authority is not relevant
Kinds of “Official” Sciences
- Exact Sciences
- Mathematics. Truth abut imaginary things.
- Positive Sciences
- what is “put” outside.
- the observer is not part of the system.
- “objective reality”.
- Natural Sciences
- Reality is the Nature.
- Social Sciences
- Reality is the Human Society.
Technology
In this framework, Technology is about Things Built by Humans
- Machines
- Processes
- Know how…
Long term Homework
Using any recording device (paper, cell phone, etc), take note of the questions that you can ask about what you see every day
Especially about questions that you don’t know the answer
For example “Does Technology derive from Science?”
Four Paradigms of Science
according to Microsoft Research
1. Empiric (since prehistoric times)

- observation of isolated facts
- description of related facts
- e.g. Botany, naming stars,
- Represended by the Arab astronomers, Galileo, Tycho Brahe, Carl Linneaus
2. Theoretical (Renaissance)

- Abstract models and theories
- Usually expressed in mathematical formulas
- Correct predictions validate the models
- e.g. Mendel laws of inheritance, Darwin natural selection theory, Kepler law of planet’s motion, Newton’s law of Gravity
3. Simulation Based (Mid 20th century)

- Models that cannot be expressed in formulas
- Formulas that cannot be solved
- e.g. Protein structure prediction, three body problem, galaxy modeling
- Computational Astronomy, Computational Biology
Represented by John Von Neumann

4. Data Based (21st century)

- Discovering patterns hidden in data
- Huge volumes of data
- Complex interactions
- e.g. Bioinformatics, Astroinformatics, Data mining
- Big Data, Machine Learning
How?
Measuring Gene Expression
More precisely, mRNA concentration
What is the question?
We want to know
- Which genes are being expressed
- How much of each gene is being expressed
- How does expression change
- In time
- Under different conditions
- Between strains/mutants/cell lines
The Big Assumption
Measuring protein concentration is hard
We assume that protein concentration is proportional to mRNA concentration
- Which genes are being transcribed
- How much of each gene is being transcribed
- How does transcription change
- In time
- Under different conditions
- Between strains/mutants/cell lines
How to measure mRNA concentration?
Basically
- qPCR
- Microarrays
- RNAseq
qPCR
If you have primers for each gene
- specific to each gene
- thermodynamically stable
- efficient
Raw data: CT value for each gene/condition
and CT value for calibration reference
Hybridization methods
Southern/Northern/Western blot can detect, but not quantify
(I think so. I’m not a biologist)
Instead, we have macro- and microarrays
Raw data: Light intensity (luminescence) in one or more wave length
This is measured in arbitrary units, and is a number between 0 and 65536
(that is, a 16-bits value)
RNAseq
mRNA is retro-transcribed and fragmented.
Fragments are sequenced. Reads are aligned to reference genome
Raw data: SAM/BAM file with location of each read in the reference genome
Processed data: Number of reads per gene, normalized by gene length
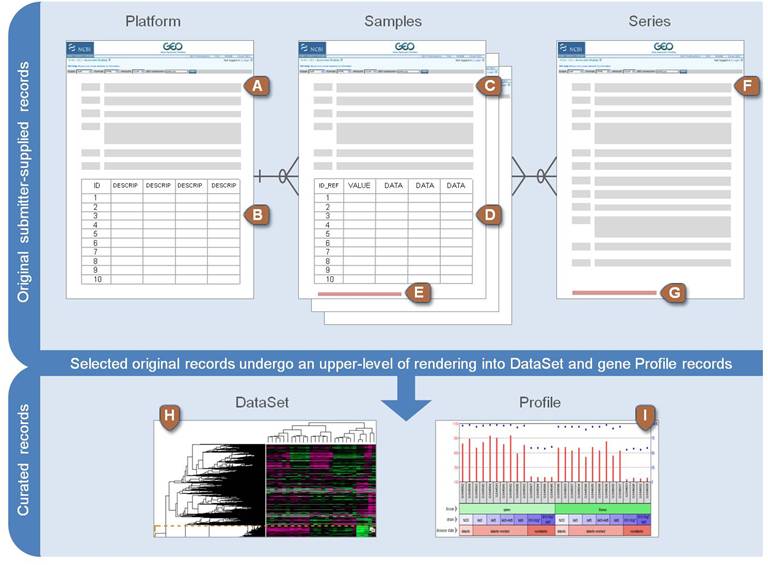
Data source: NCBI GEO
Gene Expression Omnibus
- Platforms
- Samples
- Series
- Data Set
- Profile
Relevant Objects in GEO
- GEO Platform
- Set of probes used in one or more experiment. Type of microarray slide, qPCR primers, including controls.
- GEO Samples
- a specific result of a single experiment. Raw RNA concentration for each probe in the platform
- GEO Series
- Set of Samples from a complete experiment. Includes technical and biological replicas
Relevant Objects in GEO
- GEO Datasets
- Sets of samples from different experiments that can be compared. For example, using the same platform
- GEO Profiles
- individual gene expression profiles assembled from GEO. Follows a single gene through several conditions
NCBI GEO data structure
Types of files
NCBI standard
- SOFT
- MINiML
- Series Matrix
Industry standard
- CEL (Affymetrix)
- GPR
- FASTQ (NGS)
- SAM/BAM (RNAseq)
Homework
- Learn how to read these files in your computer
- They are usually compressed
- Do not use Word
- If you use Excel, be careful
- Learn how to get data from the European Database
Write a document (in English) explaining your results